This week I had a paper published in Evolution Letters. It’s always a great feeling to get a paper through the trial-by-fire that is peer-review but this one was very special to me. The work we present was also the second chapter of my PhD thesis, and one of my favourite projects to work on during my studies, so it is very gratifying to have it finished. Here I’ll give you some background to the project and an overview of what we found.
The project started long before I became a biologist, or even started my studies. In 2002, I (see below) was 14 years old and probably more concerned with pokémon than what fruit flies are getting up to in their daily lives. Meanwhile, Rhonda Snook, who is now Professor at Stockholm University and was then at the University of Sheffield, was busy setting up an ambitious experiment. She was originally interested in understanding why males of the the fruitfly Drosophila pseudoobscura produce different types of sperm. The hypothesis was that sexual selection, the process produced by the competition between individuals over reproductive resources and mating opportunities, was involved. To directly test this idea, she set up an experimental evolution project which manipulated the sexual selection environment. In the general approach, you set up different “environments” and then letting populations of flies evolve and adapt to these environments over many generations after which you can study the evolutionary response by comparing the organisms that have been evolving in the different environments. In one treatment, elevated promiscuity, individual females were kept in tube with six males for her reproductive life, while in another treatment, enforced monogamy, a female was kept with only a single male. The main difference between the treatments was therefore the level of competition among males for the reproductive resources of the female, generating strong sexual selection in the elevated promiscuity treatment, and abolishing it in the enforced monogamy treatment. The same treatments were applied every generation, using the offspring emerging from the previous generation, with 1 generation being ~30 days. Each treatment was replicated 4 times so that maintaining each generation involved ~2000 flies that had to be fed, moved around, and maintained every month. You can read more about the results with respect to how these treatments affected the sperm morphology of males here.
 |
|---|
| Your author ca. 2002 |
 |
|---|
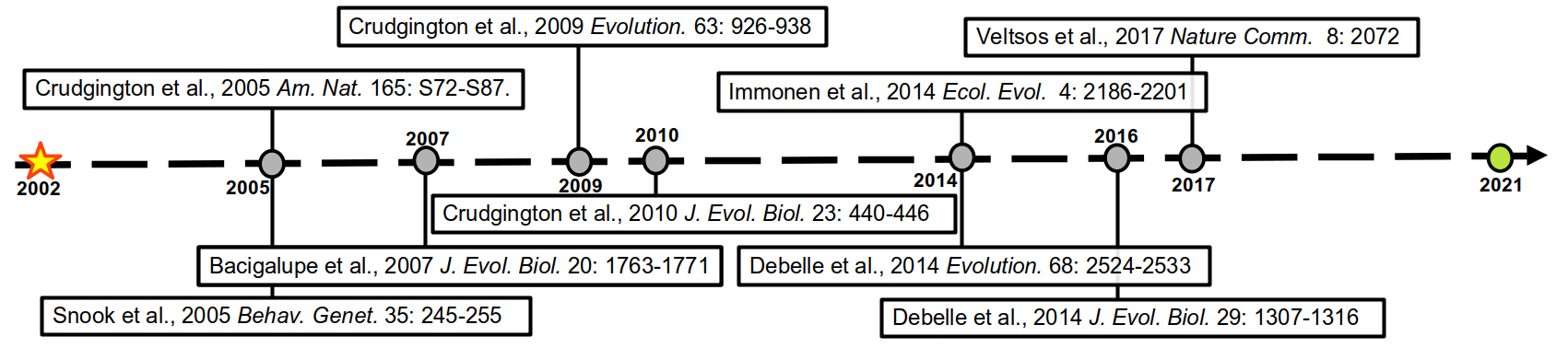
| The timeline of the project |
This experiment was kept going for many generations, and rounds of funding, with several studies investigating what other evolutionary changes to males and females these altered mating systems had produced. In figure 1 (above) is a timeline of these studies spanning a total of nearly 20 years, and counting. There are too many to summarise here, but they are full of interesting results and you should definitely look them up. This most recent publication in Evolution Letters is the green dot.
I came on the scene when in 2013, bright-eyed and enthusiastic, I started my PhD studies under the supervision of Professor Mike Ritchie at the University of St Andrews. Mike had, at that time, already collaborated with Rhonda for some time on several experiments with these experimentally evolved flies. By now the “genomics revolution” was in full-swing and whole-genome sequencing was all the rage. After finding phenotypic responses such as changes to courtship song characteristics, and more harmful males with higher courting rates in the elevated promiscuity treatment, time had come to characterise the genetic changes between the elevated promiscuity and enforced monogamy treatments. By now these flies had been evolving for ~160 generations (11-12 years). For context, humans are generally given a generation time of ~30 years (the time it takes, on average, for a human to grow, develop into an adult, and produce new offspring). With that generation time, 160 generations is equivalent to 4,800 years, which, counting back from today, would mean a start of the experiment in ~2800 BCE, around the time that the construction of the pyramids of Egypt began.
An important part of the context for this work is that researchers have been very interested in the role that sexual selection plays in evolution, especially its role in shaping genetic variation. Studies comparing the genomes of different populations or species often find differences at genes that have an important role in mating behaviours, female preferences, and male signalling. These give a tantalising hint that sexual selection is indeed an important component of population divergence and speciation, processes at the core of evolution. However, because there are so many other differences between populations and species, for example ecology, it’s very difficult to infer that a particular change was driven by selection in the context of mating (i.e. sexual selection). Experimental evolution offers an elegant solution to this problem. Because we can set up the experiment in the lab, we can control all other variables and only change the mating system or the level of sexual selection. We can then attribute genetic changes between the treatments to the variable we have changed (the level of sexual selection).
We already knew that there were differences between the elevated promiscuity and enforced monogamy lines in gene expression but what was not known is whether there were any alleles or genetic markers that had changed in frequency between the elevated promiscuity and enforced monogamy populations. So my task became to extract DNA from samples of flies from each replicate, get the genomes sequenced, and then compare the genomes of the elevated promiscuity and enforced monogamy lines to identify genetic differences between them.
This problem of identifying genetic changes, at the whole-genome scale, led me down an unexpected detour evaluating different statistical approaches and eventually propose some improvements in a separate paper in Methods in Ecology and Evolution). In the current Evolution Letters paper we show that there have indeed been many genetic changes between the two treatments. We also show that there are more differences than expected by random fluctuations in allele frequencies alone. We also show that these changes occur in quite distinct clusters in the genome (see, for example, (1) in the figure below). We were also able to show that, in many cases, the regions of the genome within these clusters show reduced genetic variation within the elevated promiscuity or enforced monogamy treatments (see (2) in the figure below for a reduction in diversity in elevated promiscuity but not enforced monogamy lines under the peak of genetic differences). Such reductions in diversity are very characteristic signals of selection within genomes. These regions also contain several interesting genes that are known to be involved in mating behaviours and other traits related to reproduction. genes that stand out include seminal fluid proteins. These are proteins that are passed to females during mating and have a variety of effects on the physiology of mated females, making them very good candidates for sexual selection. Finally, the changes between elevated promiscuity and enforced monogamy lines are over-represented on the X-chromosome, which is thought to be a “hot-spot” for genes under sexual selection.
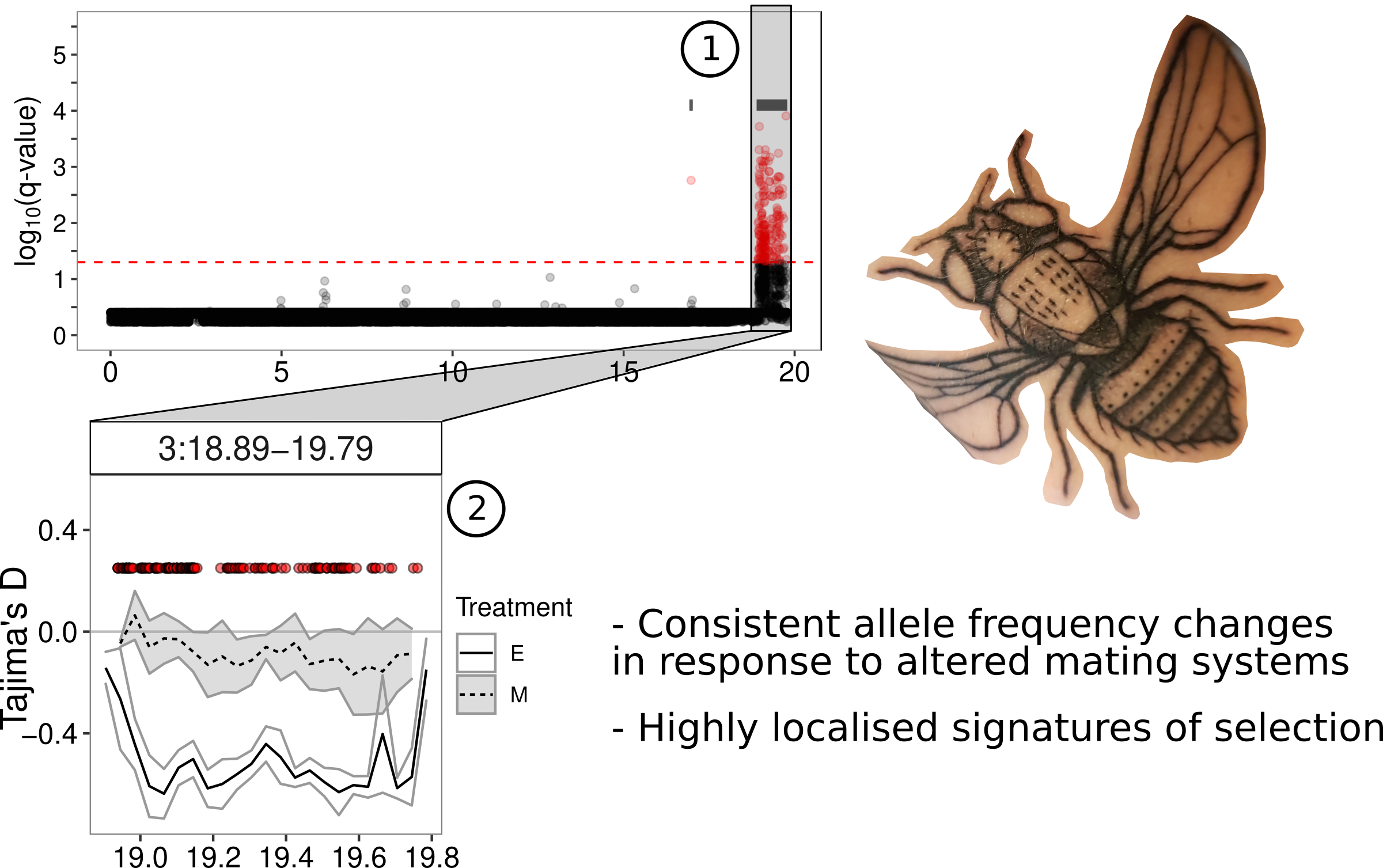 |
|---|
| A small depiction of the main findings and outcomes |
In summary, this paper is an important contribution to the field of evolutionary biology. We are able to isolate the process of sexual selection from other factors and investigate direct changes in response to this process. This has been a challenge in comparisons of natural systems where populations and species often vary in many different aspects. Fully understanding the consequences of these genetic changes will require more work but we are super excited to have taken one more step in understanding the role of sexual selection as an evolutionary force.
My co-authors in on this paper were Paris Veltsos, Mike Ritchie, and Rhonda Snook.
The full paper is open access, and you can read the paper here if you want to learn more about this. Get in touch if you have questions!
A version of this post also appears on the Evolution Letters blog Thanks to Paris, Mike, and Rhonda for helpful comments on this post.